SARS-CoV-2 is critically dependent on a special mechanism for the production of its proteins. A collaborative team led by a research group at ETH Zurich obtained molecular insights into this process and demonstrated that it can be inhibited by chemical compounds, thereby significantly reducing viral replication in infected cells.
Video: Said Sannuga, Cellscape.co.uk / ETH Zurich, The Ban Lab
Viruses require the resources of an infected cell to replicate and then infect further cells, and transfer to other individuals. One essential step in the viral life cycle is production of new viral proteins based on the instructions in the viral RNA genome. Based on these construction plans, the cell’s own protein synthesis machine, called the ribosome, produces the viral proteins.
In the absence of viral infection, the ribosome moves along the RNA in strictly defined steps, reading three letters of RNA at a time. This three-letter code defines the corresponding amino acid that is being attached to the growing protein. It almost never happens that the ribosome slips one or two RNA letters forward or backward instead of following the regular three-letter steps. When such a slip of the ribosome occurs, it is called “a frameshift,” and it leads to incorrect reading of the genetic code.
Frameshifting almost never happens in our cells. It would lead to dysfunctional cellular proteins; however, certain viruses, such as coronaviruses and HIV, depend on a frameshifting event to regulate levels of viral proteins. For example, SARS-CoV-2 - the virus that causes COVID-19 - is critically dependent on frameshifting promoted by an unusual and intricate fold in the viral RNA.
Therefore, since frameshifting is essential for the virus but it almost never happens in our organism, any compound that inhibits frameshifting by targeting this RNA fold could potentially be useful as a drug to combat infection. However, so far, there is no information on how the viral RNA interacts with the ribosome to promote frameshifting, which would be important for drug development.
Detailed image of an essential process for coronavirus replication
A team of researchers from ETH Zurich and the Universities of Bern, Lausanne and Cork (in Ireland) has for the first time managed to reveal the interactions between the viral genome and the ribosome during frameshifting. Their results have just been published in the journal Science.
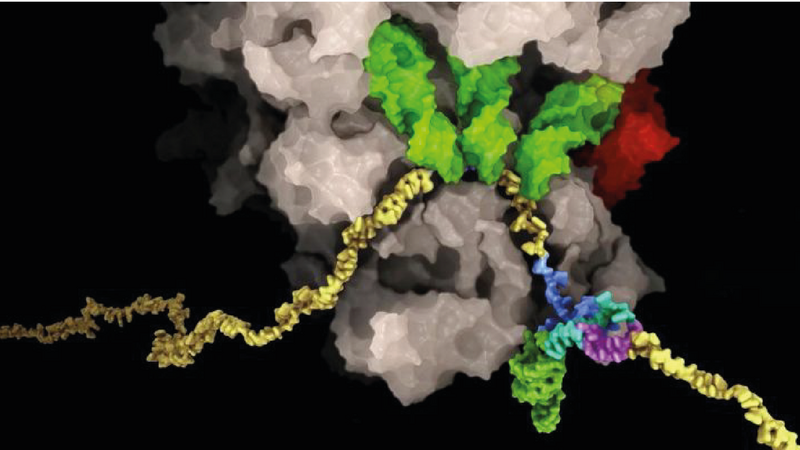
Using sophisticated biochemical experiments, the researchers managed to capture the ribosome at the frameshifting site of the SARS-CoV-2 RNA genome. They could then study this molecular complex using cryo-electron microscopy.
The results provided a molecular description of the process at unprecedented detail and revealed a number of novel unanticipated features. The frameshifting event causes the usually dynamic ribosome machine to adopt a strained conformation, which helped provide one of the sharpest and most accurate images of a mammalian ribosome, visualized in the process of frameshifting while reading the information from the viral genome. The researchers then followed up on their structural findings with in vitro and in vivo experiments including exploring how this process can be targeted with chemical compounds. Nenad Ban, Professor for Molecular Biology at ETH Zurich and co-author of the study, emphasizes that “The results presented here on SARS-CoV-2 will also be useful for understanding the frameshifting mechanisms in other RNA viruses”.
Possible target for antiviral drug development
The dependence of SARS-CoV-2 on this ribosomal frameshifting event could be used to develop antiviral drugs. Previous studies reported that several compounds are able to inhibit frameshifting in coronaviruses, however, this study now provides information about the effects of these compounds on levels of SARS-CoV-2 in infected cells.
In their experiments, both compounds reduced viral replication by three to four orders of magnitude and were not toxic for the treated cells. However, one of the two reduced viral replication by inhibiting ribosomal frameshifting, while the other might act through a different mechanism.
Although these compounds are currently not potent enough to be used as therapeutic drugs, this study demonstrates that inhibition of ribosomal frameshifting has a profound effect on viral replication, which paves the way for the development of better compounds. Due to the fact that all coronaviruses depend on this conserved frameshifting mechanism, a drug that targets this process may even be useful to treat infections by more distantly related coronaviruses. “Our future work will focus on understanding the cellular defense mechanisms that suppress viral frameshifting, as this could be useful for development of small compounds with similar activity,” says Ban.
Pramod R. Bhatt, Alain Scaiola, Gary Loughran, Marc Leibundgut, Annika Kratzel, Romane Meurs, René Dreos, Kate M. O'Connor, Angus McMillan, Jeffrey W. Bode, Volker Thiel, David Gatfiled, John F. Atkins, Nenad Ban: Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science; published online 13 May 2021
https://science.sciencemag.org/content/early/2021/05/12/science.abf3546