The coding parts are called exons, the non-coding sequences are called introns. The information of a gene is therefore fragmented and must be assembled during gene expression. In order to do this, an RNA copy of the gene is produced as a precursor: the pre-mRNA (precursor mRNA). The introns of this pre-mRNA are then recognized and cut out by a huge molecular machine, the spliceosome. This process is called splicing. After splicing and other maturation processes, the "mature" mRNA is present.
At first glance, the fragmentation of genes seems complicated and confronts the cell with the challenge of identifying and eliminating the non-coding parts without error. Why was this complex architecture favored by evolution?
The modular structure of genes offers the advantage that the information-carrying sequences can be joined together in different combinations. This process is also called alternative splicing. Humans can produce more than 100,000 different gene products from around 22,000 genes. As alternative splicing is more common in more complex organisms, it is speculated whether this mechanism has played a central role in the development of higher order organisms.
An exon between life and death
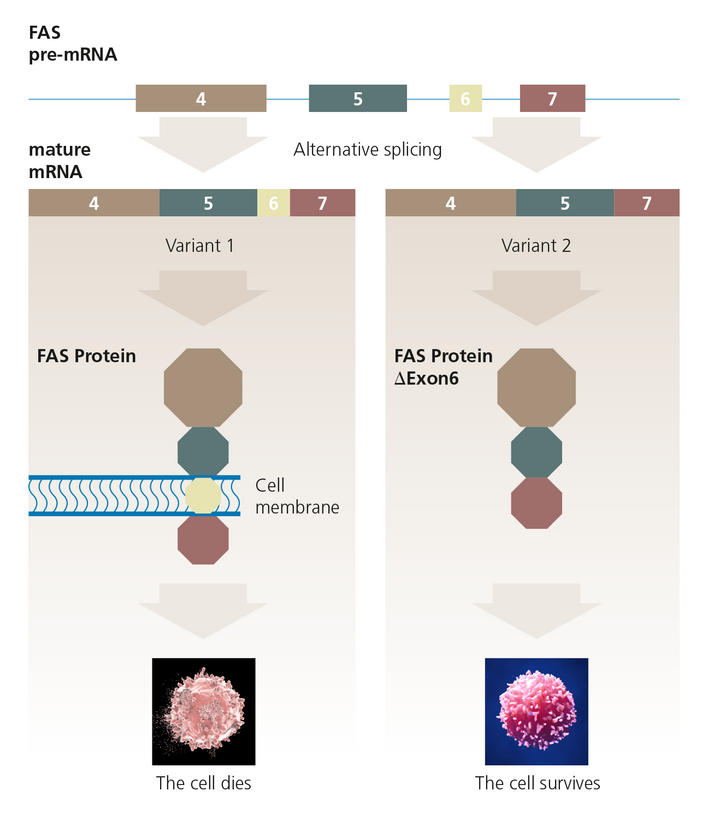
Even small differences in the splicing pattern of a gene can have dramatic effects. In the example shown here, the presence or absence of exon 6 in the mature mRNA of the FAS gene determines whether the protein produced, promotes cell survival or, on the contrary, leads to its death. This strictly regulated form of cell death is a useful way to intentionally exit under certain circumstances and serves the well-being of the organism. It was discovered early on that mutations that influence gene splicing are responsible for diseases. Examples are cystic fibrosis, spinal muscular atrophy, Duchenne muscular dystrophy and various cancers.